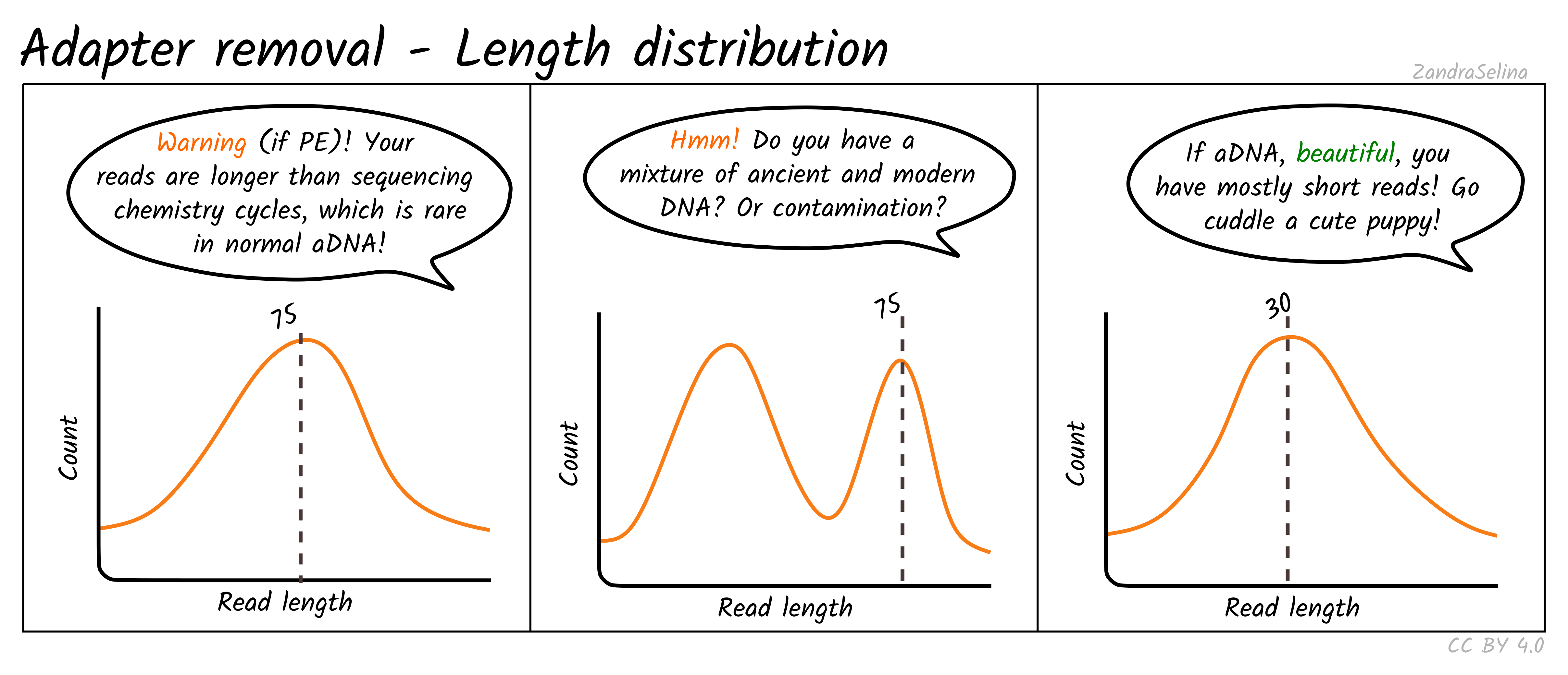
paired end sequencing read length
On sequencing using unpaired reads shows that ultra-short reads theoretically allow whole genome re-sequencing and de novo assembly of only small. Read length Sanger sequencing 500 to 600 bp Max up to 900bp Maxam Gilbert sequencing 500 to 600bp Sequencing by synthesis.
Leaving it at 25bp and having read length the splicing mapping is basically worthless.
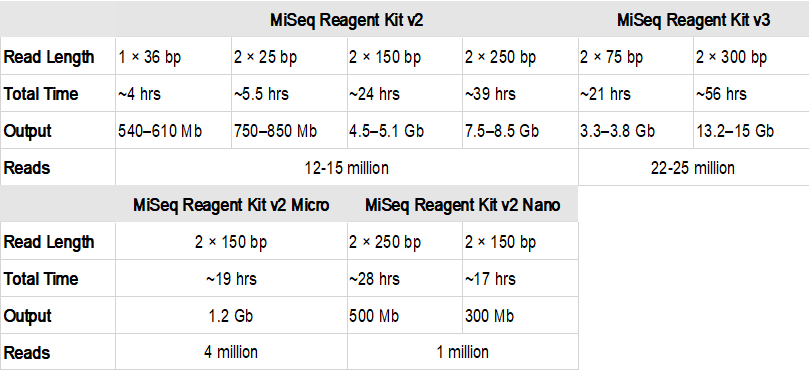
. An analysis by Whiteford et al. The paired-end short read lengths are always 2 x 150bp 300bp. Many platforms Illumina Genome Analyzer Applied Biosystems SOLID Helicos.
Next-generation sequencing technology is enabling massive production of high-quality paired-end reads. Paired-end sequencing allows users to sequence both ends of a fragment and generate high-quality alignable sequence data. To ensure sequencing quality of the Index Read do not exceed the supported read.
We use an Illumina MiniSeq for our short-read sequencing runs. You can set that 25bp to be 20 or. Many platforms Illumina Genome Analyzer Applied Biosystems.
Next-generation sequencing technology is enabling massive production of high-quality paired-end reads. Enter up to 20 characters or use manual mode if you need between 20 and 100 bp. Modern nextgeneration sequencing platforms offer a range of read configurations such as single-read SR and paired-end PE sequencing with 75 bp per read 100 bp per read and 150 bp per.
It is shown that re-sequencing and de novo sequencing of the majority of a bacterial genome is possible with read lengths of 2030 nt and that reads of 50 nt can provide. Contig length metrics were positively correlated with both read length and sequence coverage Fig. Many platforms Illumina Genome Analyzer Applied Biosystems.
The maximum distance x for a pair considered to be properly paired SAM flag 02 is calculated by solving equation Phix-musigmaxLp0 where mu is the mean sigma. When using RNA-Seq to study gene expression read length is not a significant factor. If you only have one fragment worth ie.
Whether you align 100bp paired reads or a 50bp single. Paired-end sequencing facilitates detection of genomic. What matters is read counts.
The library prep protocols are designed to. The current read length that is standard for many experiments is paired-end 100 bp reads and there is also the possibility of running paired-end 300 bp reads. 100 paired end Pyrosequencing 1000bp.
1b with the highest contig N50 2454 Mb and the longest contig. Next-generation sequencing technology is enabling massive production of high-quality paired-end reads.

Rna Extraction Method Read Length And Sequencing Layout Single End Versus Paired End Contribute Strongly To Variatio Interactive Notebooks Method Sequencing
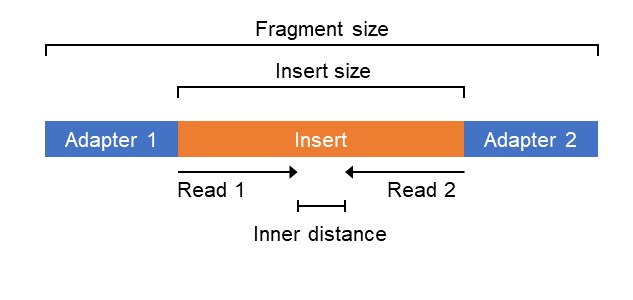
Compbio 020 Reads Fragments And Inserts What You Need To Know For Understanding Your Sequencing Data Bad Grammar Good Syntax

Beyond The Linear Genome Paired End Sequencing As A Biophysical Tool Trends In Cell Biology

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download High Resolution Scientific Diagram

Range Of Ngs Applications Rises Quickly
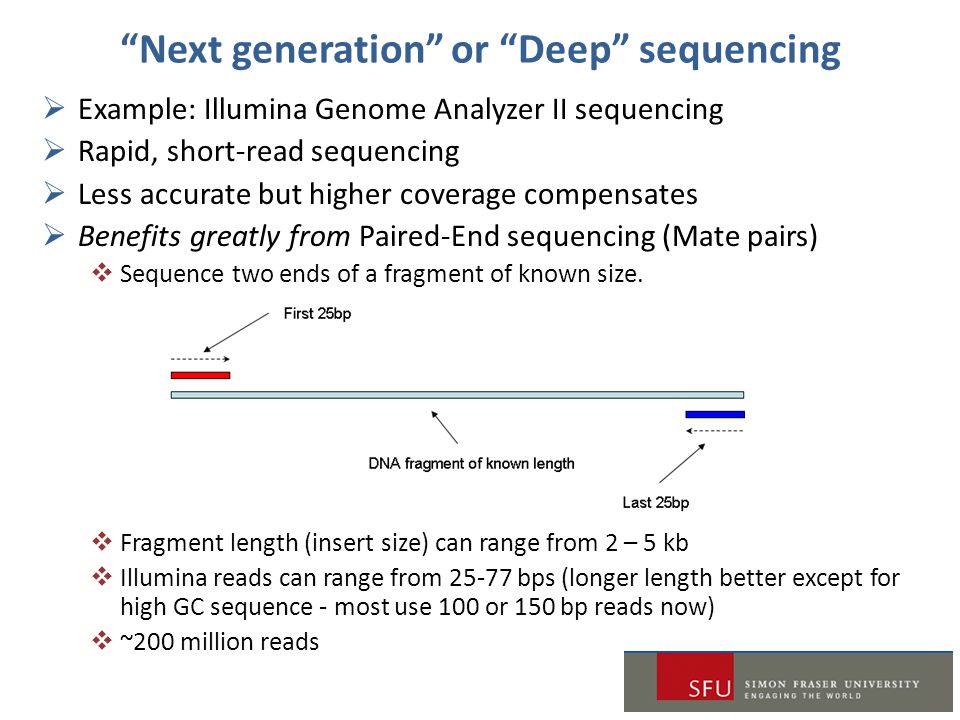
Ngs Bioinformatics Workshop 2 Ppt Download

Paired End Sequencing Left Showing Read 1 And Read 2 Primers Starting Download Scientific Diagram
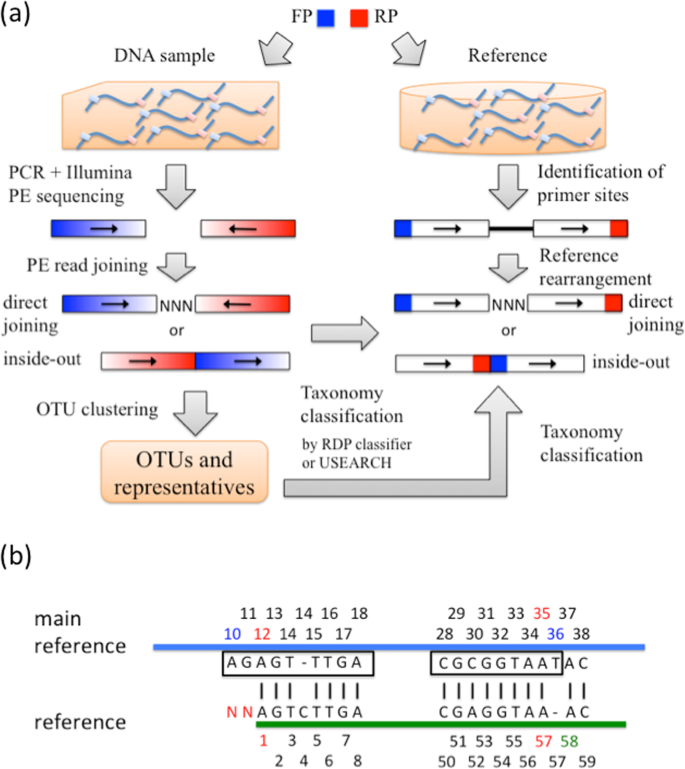
Joining Illumina Paired End Reads For Classifying Phylogenetic Marker Sequences Bmc Bioinformatics Full Text
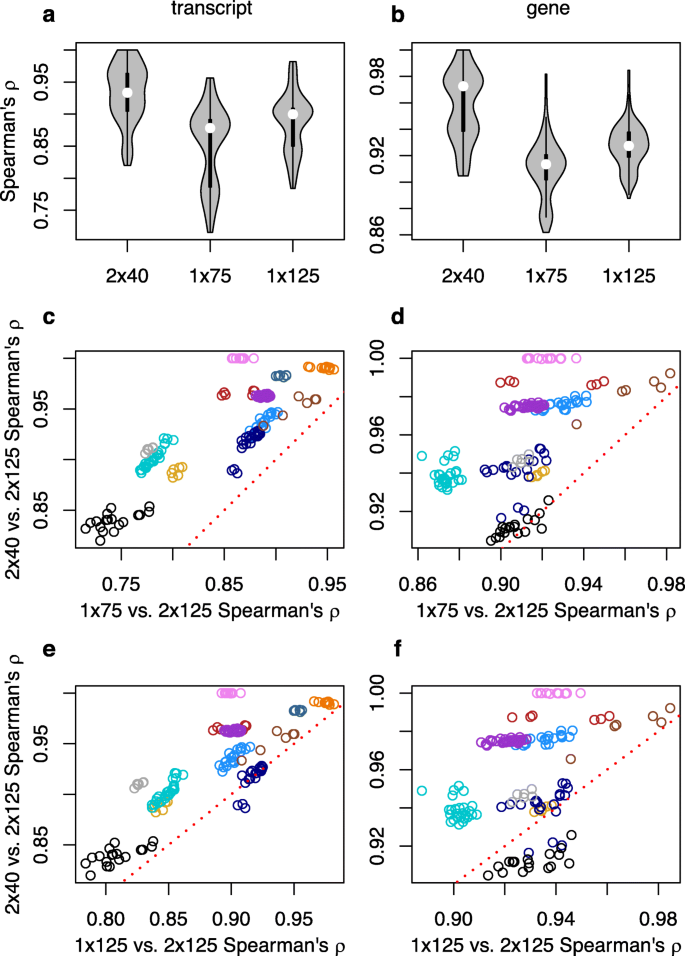
Short Paired End Reads Trump Long Single End Reads For Expression Analysis Bmc Bioinformatics Full Text
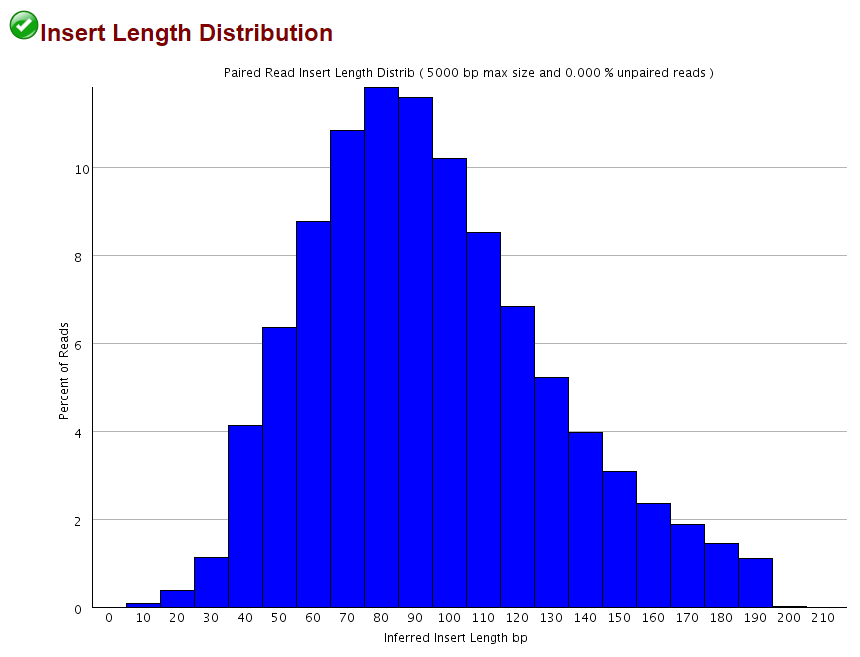
Qc Fail Sequencing Mispriming In Pbat Libraries Causes Methylation Bias And Poor Mapping Efficiencies
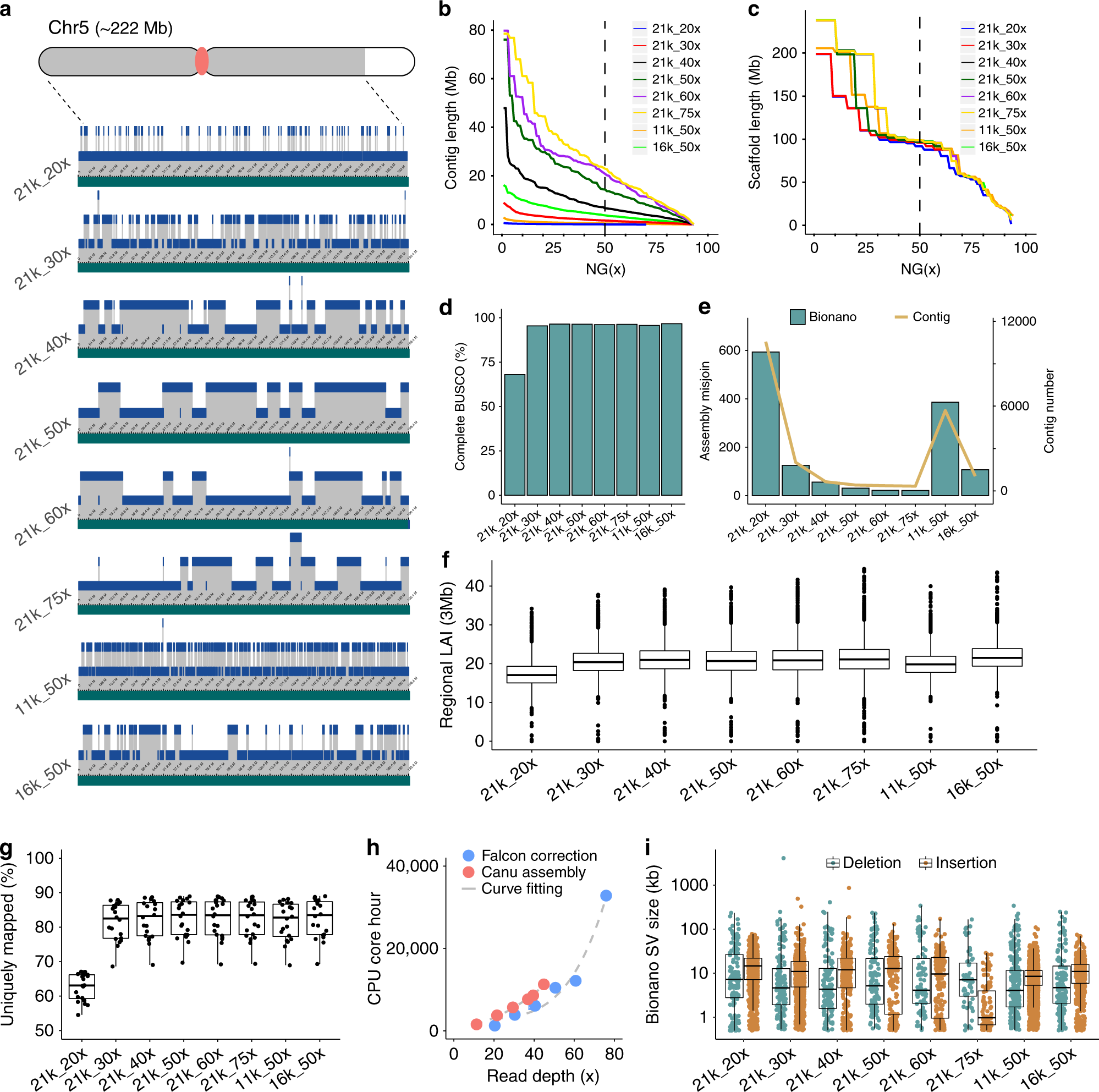
Effect Of Sequence Depth And Length In Long Read Assembly Of The Maize Inbred Nc358 Nature Communications

Comparison Of Next Generation Sequencing Systems
Local De Novo Assembly Of Rad Paired End Contigs Using Short Sequencing Reads Plos One
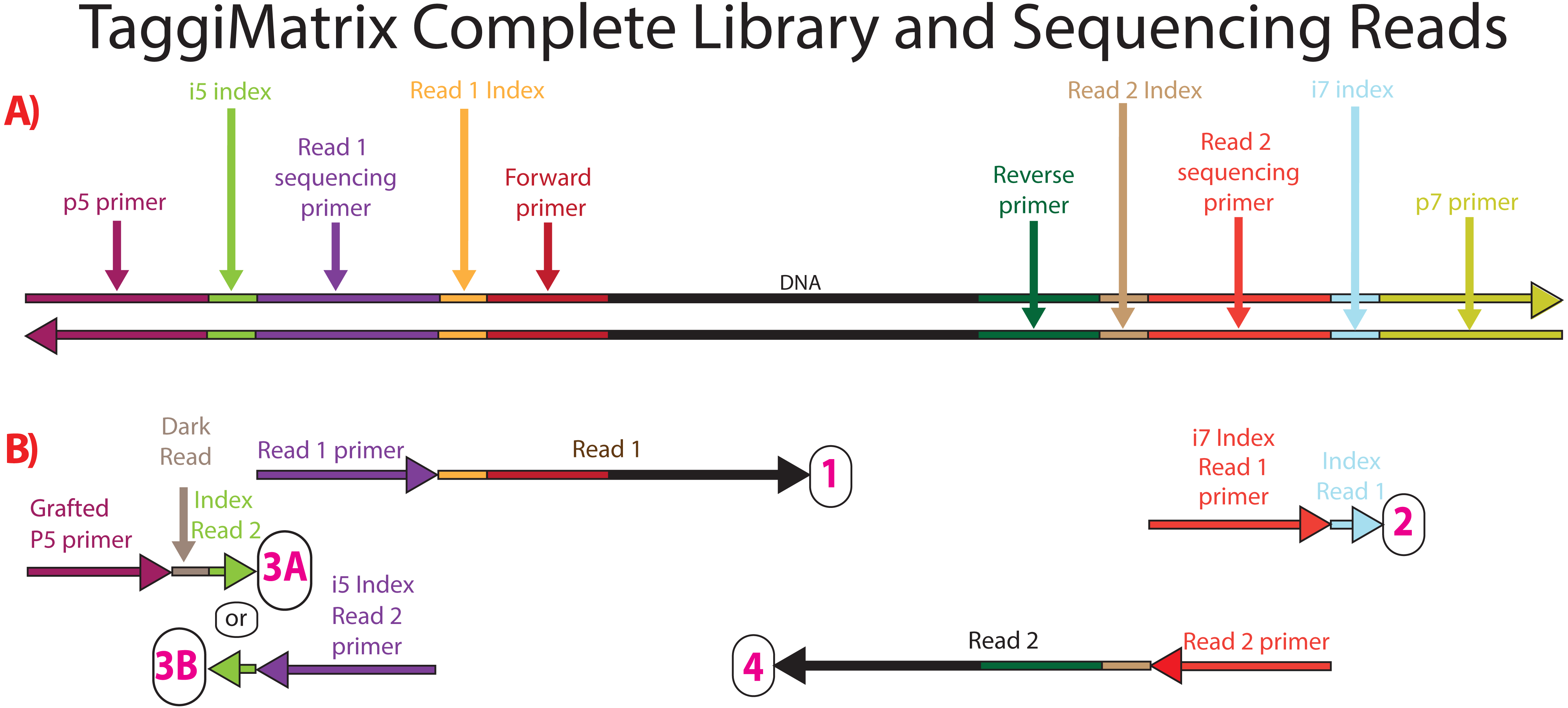
Adapterama Ii Universal Amplicon Sequencing On Illumina Platforms Taggimatrix Peerj

Interpreting Color By Insert Size Integrative Genomics Viewer